Rose Lab Research Projects
Link directly to specific projects:
- Overview
- Plant cuticle composition, archetecture and function
- High resolution functional genomics of tomato fruit development and environmental responses
- Evolution of plant cell structural polymers in Charophycean Green Algae: the immediate ancestors of land plants
Overview
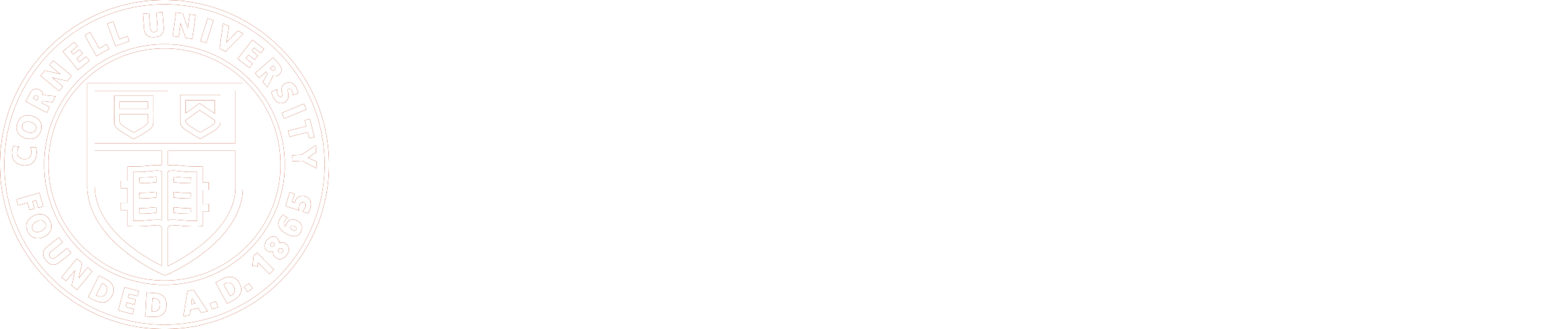
The outer surfaces of plant cells are comprised of diverse sets of polymers, including cell wall polysaccharides and proteins, as well as phenylpropanoid and lipid polymers in the case of more specialized cell types. These polymers are synthesized, secreted and assembled into elaborate matrices with architectures that vary, down to the level of heterogeneity within the specific cell wall faces of individual cells.
Cell wall composition and organization are often dynamic and can change substantially during cell growth and differentiation, or in response to environmental conditions. Research in the Rose lab addresses questions related to how this heterogeneity is established, and examines a range of biological processes at the cell surface, and in the interface between plants and their environments. The cell wall and cuticle play critical roles in providing biomechanical support, and protecting plants against pathogens and abiotic stresses. However, the relative contributions of specific compounds that are important for these different functions, and the mechanisms by which they are distributed and interact with each other within the wall framework, are not well understood.
Similarly, the biochemical environments of the apoplast (extracellular region), from the hydrophilic polysaccharide cell wall, to the hydrophobic cuticle, are likely to be highly varied in their composition with respect to the populations of resident proteins, peptide, secondary compounds and other molecules. However, this has yet to be resolved.
Here are examples of some ongoing projects in the Rose lab:
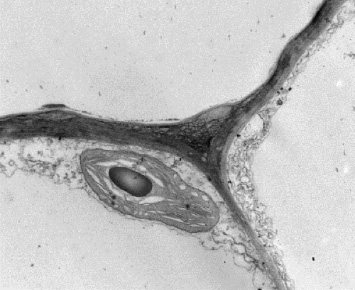
Plant Cuticle Composition, Architecture and Function
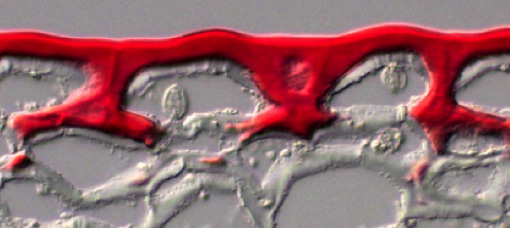
The plant cuticle is an extracellular hydrophobic layer that covers the aerial epidermis of all land plants, providing protection against desiccation and external environmental stresses. Considerable progress has been made in understanding the biosynthesis of two major cuticle components, the polymer cutin and cuticular waxes. However, the mechanisms of cuticle assembly, modification and the various functions of different cuticle components are still mostly mysterious.
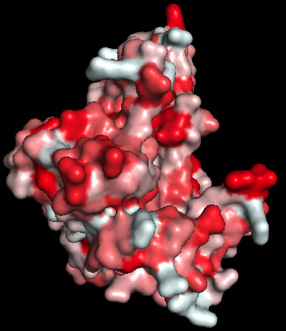
As an example, while the cuticle provides the primary barrier to water loss, remarkably little is known about the specific constituents that determine cuticle permeability, or how their synthesis is regulated. In a USDA funded project we are using tomato fruit as a model to identify the specific cuticular wax compounds that limiting desiccation.
This involves developing a detailed understanding of wax diversity, compositional dynamics and functional significance during development in response to drought stress in domesticated tomato (Solanum lycopersicum) and two wild relatives.
Specific objectives of the project include: (A) Identifying temporal and qualitative patterns of wax accumulation in cultivated tomato and wild tomato relatives (B) Characterizing the epidermal specific transcriptomes of these fruits to identify structural and regulatory gene networks that mediate cuticular wax production and deposition; (C) Using introgression lines (ILs) and backcrossed inbred lines (BILs) derived from wild tomato species to identify key genes that underlie qualitative and quantitative differences in fruit cuticular waxes; and (D) Evaluating the roles of putative wax associated structural and/or regulatory genes on wax production and their effects on cuticle properties and fruit traits.
In addition to waxes, the lab is interested in understanding the mechanisms by which the polyester cutin is assembled and modified. Specifically, we have been studying a family of proteins called cutin synthases, examining their mode of action, substrate specificity and the functions of different isoforms.
High Resolution Functional Genomics of Tomato Fruit Development and Environmental Responses

Fruit represent unique plant developmental systems and are important components of human and animal diets. Tomato, an economically important crop, has emerged as the principal model for studies of fleshy fruit development, ripening, shelf life and nutritional quality.
However, many fundamental questions related to key aspects of the development, physiology, quality traits and environmental responses of fleshy fruit remain unanswered. The Rose lab, together with Catala, Giovannoni, Fei and Mueller groups at Cornell and the Boyce Thompson Institute, are collaborating on an NSF Plant Genome Research Program project to functionally evaluate, at an unprecedented level of temporal and spatial resolution, the hierarchy of regulatory, hormonal and structural systems that influence tomato fruit ontogeny.
Another goal is to characterize poorly understood responses of fruit to drought, an increasing problem in crop production. These studies will also include two phenotypically diverse wild relatives of tomato that show a high tolerance of water stress, and so may represent opportunities for crop improvement.
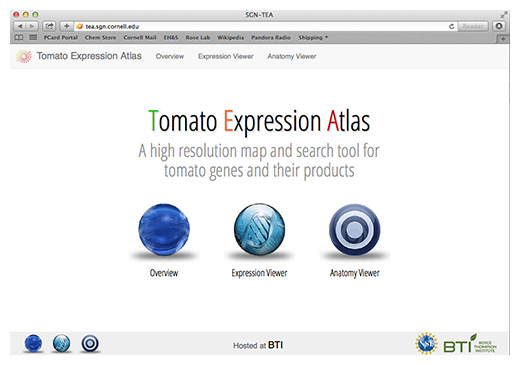
We are using laser capture microdissection (LCM) coupled with RNA-Seq based transcriptome profiling to interrogate the full range of tomato fruit pericarp cell types, as well as other tomato fruit tissues. The data are incorporated into a public ‘Tomato Expression Atlas’ (TEA) database, which is designed to display gene expression and co-expression data at the cell/tissue-type scale of resolution in multiple dimensions. The database also houses 2D, 3D and 4D images of the fruit that we are studying
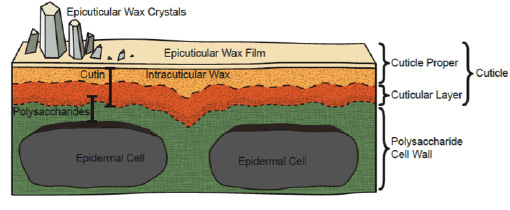
Evolution of Plant Cell Structural Polymers in the Charophycean Green Algae: the Immediate Ancestors of Land Plants
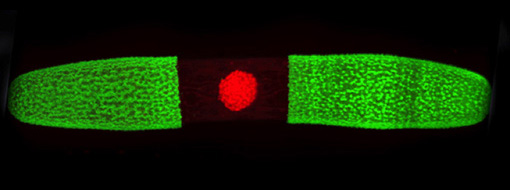
The nucleus fluoresces red.
In addition to studying the macromolecular architecture of land plant cell walls, we are also interested in their evolutionary origins. Land plants, or embryophytes, diverged from freshwater charophycean green algae (CGA) ~ 600 million years ago and subsequently radiated across essentially all the Earth’s terrestrial ecosystems. The first land plants would have needed a range of adaptations, including structural polymer networks to provide biomechanical support in the absence of a buoyant aquatic environment, and mechanisms to tolerate abiotic stresses, such damaging UV radiation and desiccation.

We are investigating the specific molecular mechanisms and evolutionary adaptations that facilitate land colonization by studying the Zygnematophyceae, the CGA lineage that comprises the closest relatives to land plants. These algae have relatively simple body plans and experience periodic drying in their semi-terrestrial habitats, which include ephemeral pools.
A critical feature of life on land is a protective and structurally complex cell wall, as well as barriers and mechanisms to resist desiccation. Together with David Domozych at Skidmore College we are using a range of CGA species to investigate the evolutionary origins of land plant cuticles and specific cell wall polysaccharides. In addition, we are developing Penium margaritaceum, a desmid in the Zygnematophyceae, as a model experimental system. This includes genome sequencing and establishing a functional genomic toolbox, including a genetic transformation protocol. We hope to use information gained through this study to better understand how plants can tolerate periodic desiccation and ultimately to improve water stress tolerance in crops.